Nature Subjournal | Improving Protein Folding Mode Coverage with Global Geometric Perception in Diffusion Models by Tsinghua and Collaborators}
Researchers from Tsinghua and other institutions enhance protein folding generation by integrating global geometric perception into diffusion models, advancing structural coverage and interpretability.


Recent breakthroughs in diffusion-based protein design have significantly advanced the field. However, evidence shows that models still struggle to generate certain fold categories, and current metrics like designability, novelty, and diversity do not fully reflect the coverage of natural protein space.
To address this, a research team from Tsinghua University, Beijing Institute of Life Sciences, and other institutions proposed an unsupervised system — TopoDiff — which learns and utilizes global geometric perception in latent representations to enable both unconditional and controllable protein generation. The study was published in Nature Machine Intelligence.

Enhancing protein fold coverage
To improve the coverage of specific protein folds, methods such as residue-level 1D/2D fold constraints combined with fine-tuning are used to generate immunoglobulin domains with variable loops or train classifiers on specific protein classes. While these methods enhance coverage, they require clear class definitions and sufficient training samples.
Therefore, the team focused on a universal unsupervised learning problem: how to train diffusion models on arbitrary datasets without relying on labels or prior knowledge to capture the underlying data distribution? Can the design advantages of SOTA generative models be leveraged to learn interpretable latent codes for deeper understanding?
Based on this, the team proposed TopoDiff, which jointly trains a diffusion-based structural generator and a structure encoder-decoder architecture to achieve two goals:
- Learn a compact, fixed-length continuous latent space encoding high-level global geometric features of proteins;
- Build a generation module that operates on residue-level features conditioned on latent information.
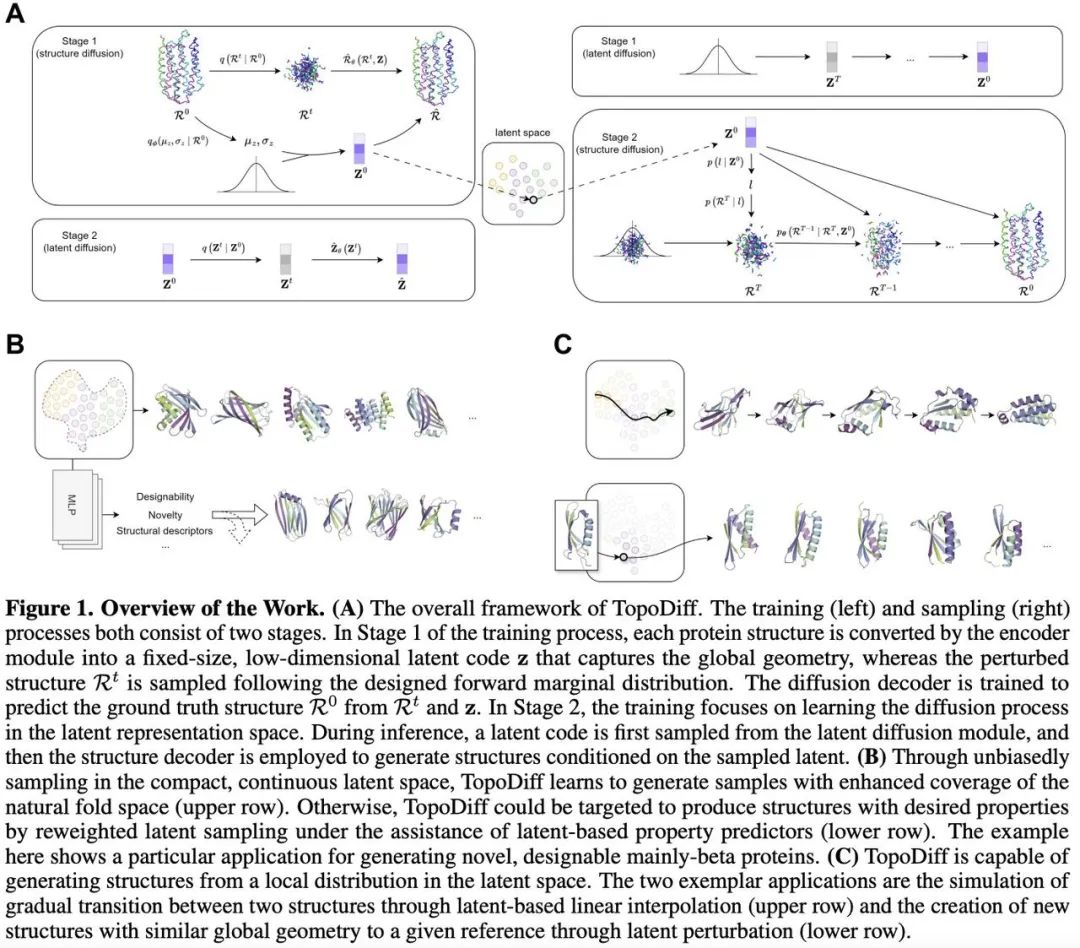
This study trained a simple latent diffusion model to unbiasedly sample learned latent distributions, guiding protein structure sampling — effectively broadening fold mode coverage and opening new dimensions for controllable generation.
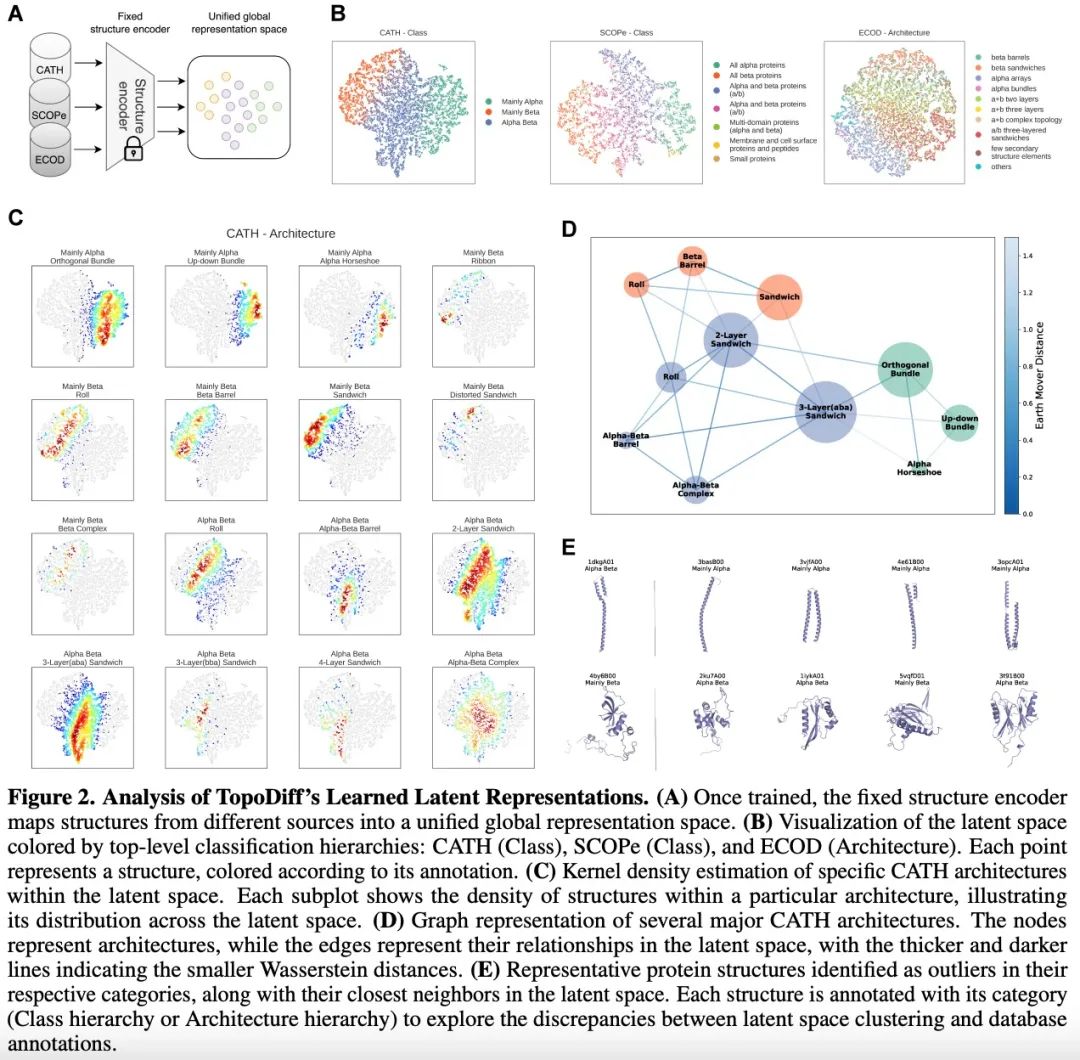
Evaluation
To assess TopoDiff’s ability to generate new protein structures unconditionally, the study compared it with several cutting-edge diffusion models, including Genie, FrameDiff, Chroma, and RFDiffusion.
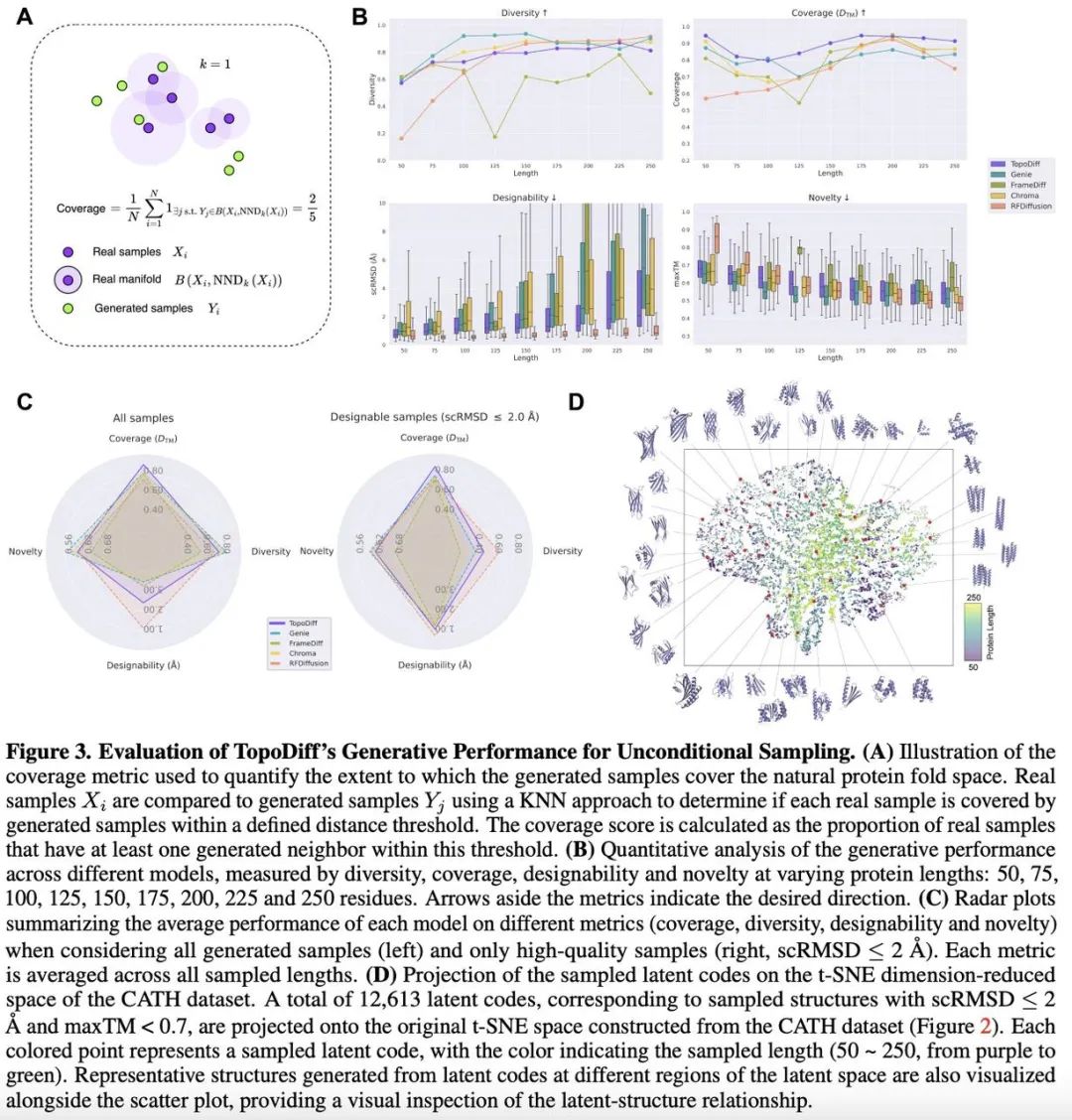
The team noted that existing evaluation metrics mainly focus on single-sample quality (like designability, novelty) or intra-sample diversity, but lack quantification of how well generated samples cover known fold space.
Coverage degree objectively reflects the model’s unbiased sampling ability within the existing data. Evidence suggests ignoring this metric can lead to biased model selection.
Over the past decade, de novo protein design has been limited to α-helix bundles and α-β sandwich structures, and diffusion models have yet to effectively reverse this trend.
Therefore, the team defined a coverage metric and systematically evaluated TopoDiff and other state-of-the-art models using this and traditional metrics like designability, novelty, and diversity.
Biological experiments confirmed that TopoDiff significantly improved the coverage of natural protein folds.
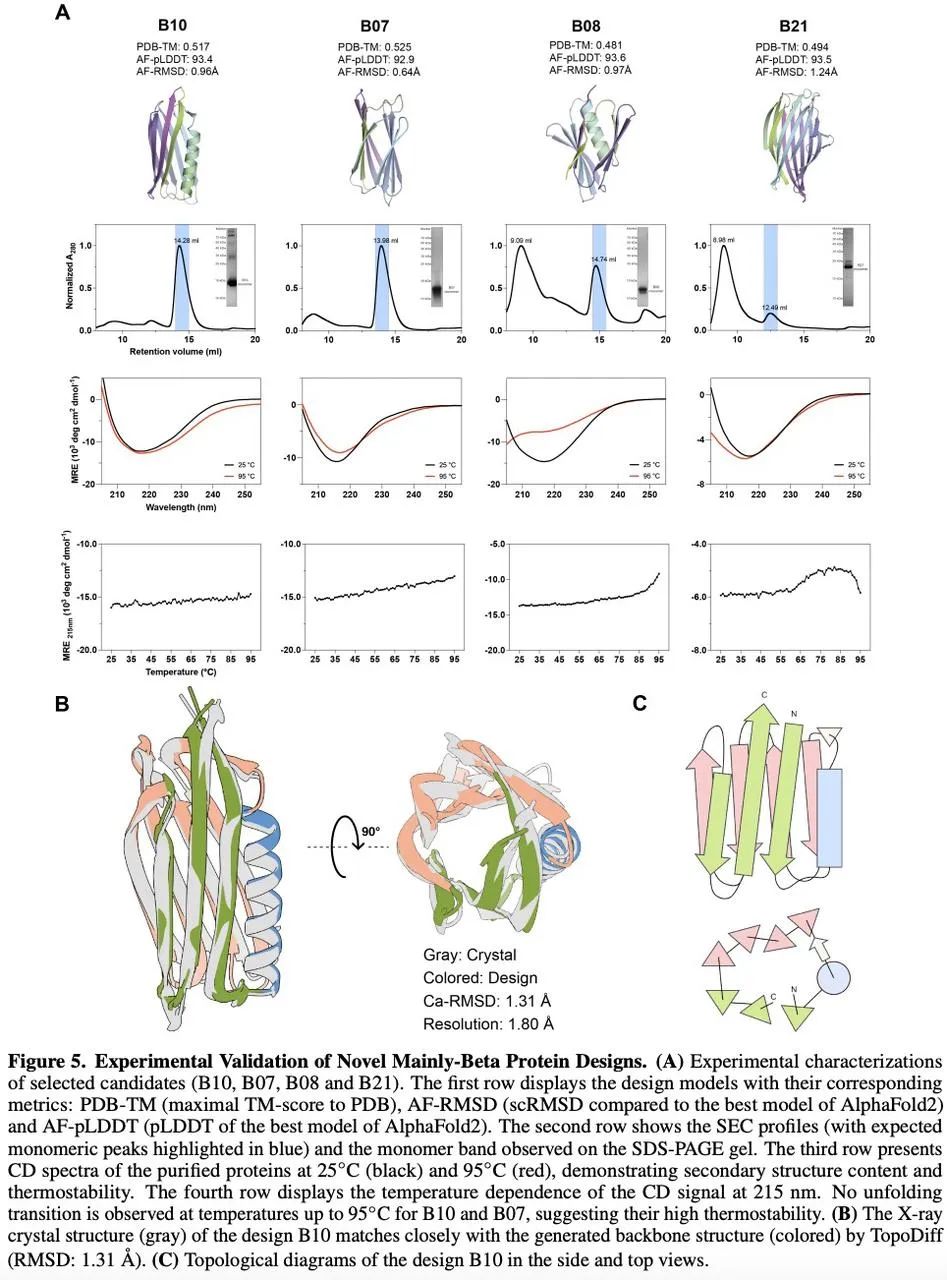
Interested readers can explore the full paper for more details on this research.