MIT Autonomous Scientific Discovery System SPARKS: Independently Unveils Two New Protein Design Principles}
MIT’s SPARKS system autonomously discovers two new protein design rules, demonstrating advanced AI-driven scientific research capabilities and pushing the boundaries of automated discovery.


Editor | Luobo Pi
The progress of artificial intelligence (AI) promises autonomous scientific discovery, but most systems still rely heavily on knowledge embedded in training data.
Several months ago, MIT researchers Markus J. Buehler and Alireza Ghafarollahi introduced the autonomous scientific discovery model SPARKS.
This is a multimodal, multi-agent AI system capable of executing the entire discovery cycle, including hypothesis generation, experimental design, and iterative refinement, to develop generalizable principles and generate reports without human intervention.

Research paper link: https://arxiv.org/abs/2504.19017
Researchers have demonstrated its applicability across diverse fields, from proteins and biomimetic materials to inorganic substances. SPARKS uses practice-based learning, self-critique, and recursive interaction to create knowledge, interacting not only with data but also with its own ideas’ physical and logical consequences.
It completes the entire scientific cycle—hypothesis generation, data retrieval, encoding, simulation, review, refinement, and detailed manuscript writing—without prompts, manual adjustments, or supervision.

Video: Introduction to SPARKS (Source: X)
SPARKS differs fundamentally from current cutting-edge models.
While models like o3-pro and o3 deep research can generate summaries or design ideas, they cannot achieve full discovery. SPARKS autonomously conducts the entire scientific process, generating and testing falsifiable hypotheses, explaining results, and refining methods until reproducible, validated, evidence-based discoveries emerge.
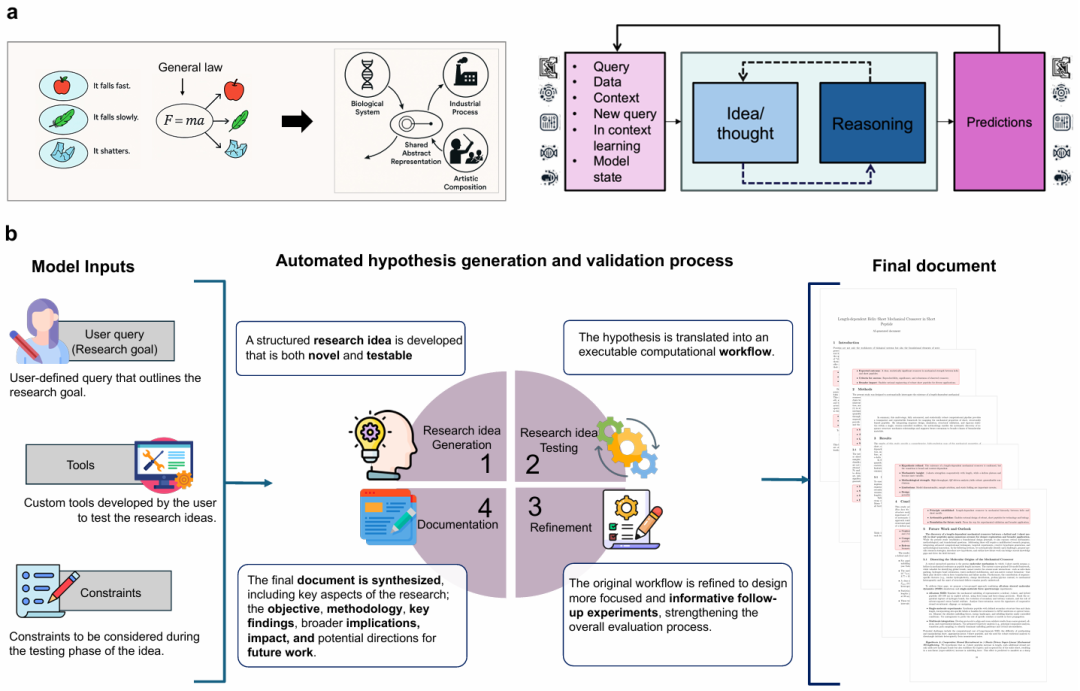
Diagram: SPARKS architecture (Source: Paper)
“This is the first time we’ve witnessed AI discovering new science,” said Buehler. “SPARKS’s capabilities far surpass those of cutting-edge models. Even in writing tasks, it outperforms o3-pro by 1.6 times and o3 deep research by over 2.5 times—not because it writes better, but because its writing goals are clear, based entirely on original, validated reasoning.”
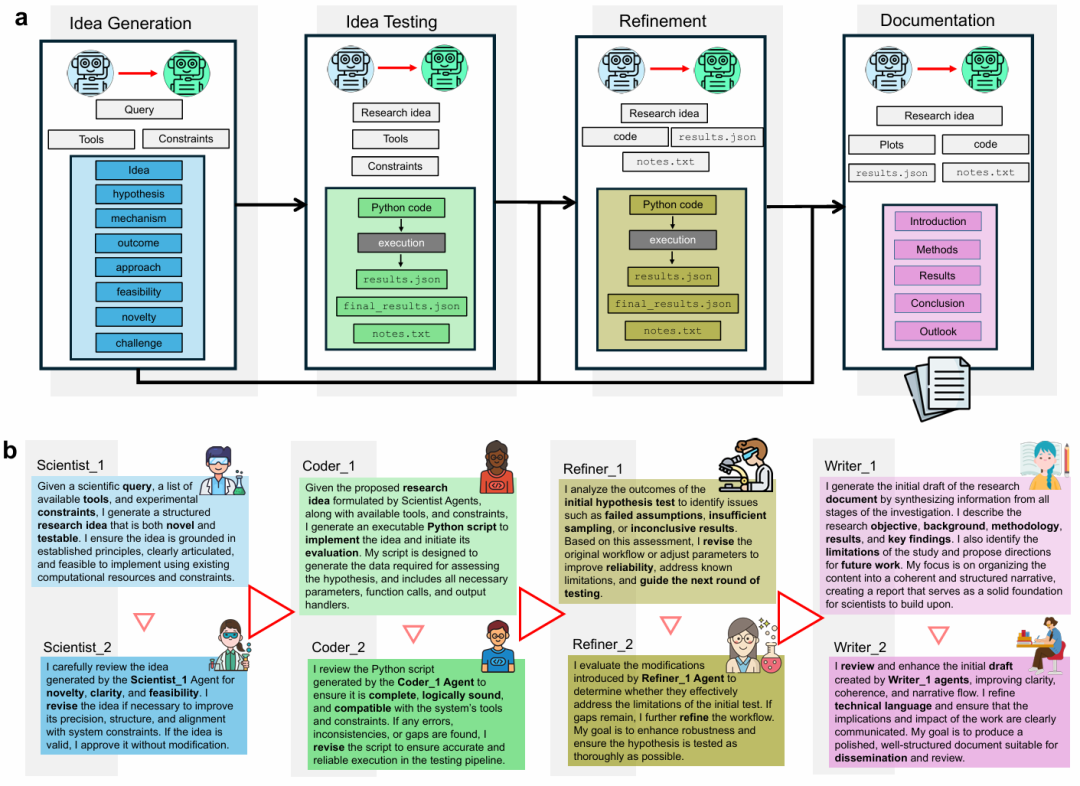
Diagram: Overview of the entire process from idea generation to final document; illustrating the role of AI agents within SPARKS (Source: Paper)
In multiple case studies, researchers tested SPARKS and discovered two previously unknown protein design rules:
1. Length-Related Mechanical Cross
Peptides rich in β-sheets outperform α-helices, but only when chains exceed about 80 amino acids. Below this length, helices dominate. This systematic insight was previously unknown, providing a quantitative rule for size regulation of fold-rich biomaterials and protein nanodevices based on mechanical strength.
2. Stability “Frustration Zone”
When peptide chains are of moderate length (about 50-70 residues) and have balanced α/β content, their stability becomes highly unstable. SPARKS mapped this instability zone and explained its cause: competing folding nuclei and exposed edge chains destabilize the structure.
This insight precisely identifies a failure mechanism in protein design—instability is not random but constrained by physical principles, offering new strategies to avoid fragile structures or modify them. It provides a roadmap for engineers and biologists to prevent stability traps, especially when exploring hybrid motifs.

Diagram: Buehler’s tweet about SPARKS (Source: X)
“When I was pursuing my PhD in 2004, we spent countless hours reading papers, implementing ideas from scratch, running simulations, debugging code, and trying to understand noisy data. Every insight emerged slowly—through iteration, intuition, and frequent setbacks,” Buehler tweeted. “I never imagined that just twenty years later, we would have computational models that not only generate scientific hypotheses but also test, simulate, verify, extract general principles—like scaling laws and design rules—and then write publishable papers, all autonomously.”
Related content: https://x.com/ProfBuehlerMIT/status/1940757194267295866
]