Microsoft Research BioEmu Published in Science: Redefining Protein Function Research with Generative AI}
Microsoft's BioEmu leverages generative AI to simulate protein conformations with unprecedented efficiency, advancing understanding of protein functions and accelerating drug discovery.


On July 10, Microsoft Research AI for Science team published their latest research in Science titled “Scalable emulation of protein equilibrium ensembles with generative deep learning”.
- Paper: https://www.science.org/doi/10.1126/science.adv9817
- Code: github.com/microsoft/bioemu
- Model: https://huggingface.co/microsoft/bioemu
- Benchmark: github.com/microsoft/bioemu-benchmarks
- ColabFold: https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/BioEmu.ipynb
- AI Foundry: https://ai.azure.com/catalog/models/BioEmu
Researchers introduced BioEmu, a generative deep learning model capable of simulating protein conformational changes with unprecedented efficiency and accuracy, opening new avenues in understanding protein functions and drug discovery.
From Structure Prediction to Functional Simulation: The Next Frontier in Protein Research
While models like AlphaFold have made breakthroughs in predicting static protein structures, they struggle to capture the dynamic conformational changes essential for understanding protein functions. Proteins are not static; they exist as ensembles of conformations, and their functions depend on transitions between these states.
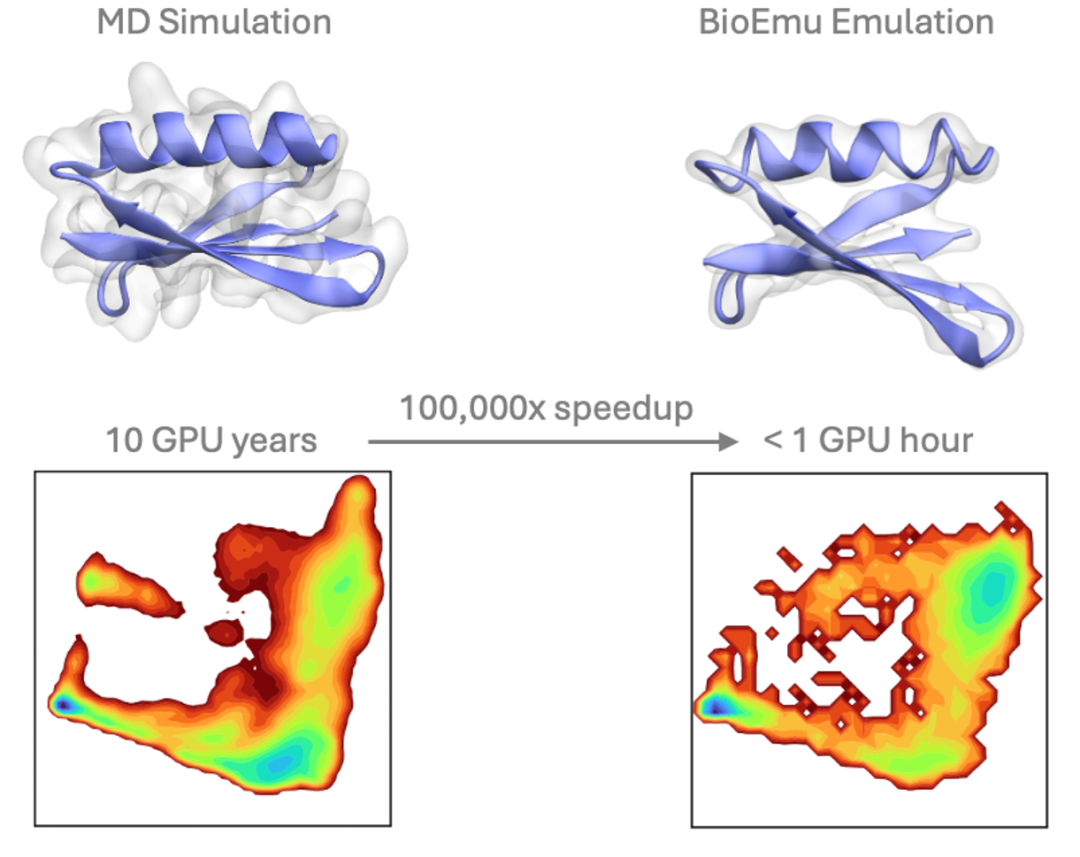
BioEmu was developed to address this challenge. It combines static structures from AlphaFold, over 200 milliseconds of molecular dynamics (MD) simulation data, and 500,000 experimental data points on protein stability. It can generate thousands of independent protein conformations per hour on a single GPU.
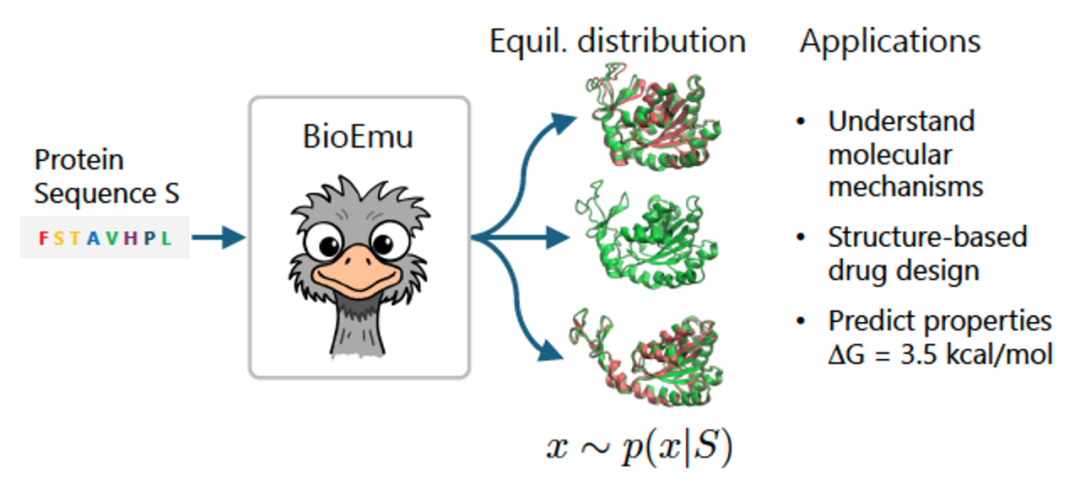
Video: Protein conformations generated by BioEmu
Generative Modeling
BioEmu builds on Microsoft Research’s previous work DiG (Distributional Graphormer), using a diffusion model architecture combined with AlphaFold’s evoformer encoder and second-order sampling techniques to efficiently sample from protein conformational distributions. Its core innovations include:
- Simulating key structural changes during protein function, such as cryptic pockets, local unfolding, and domain rearrangements;
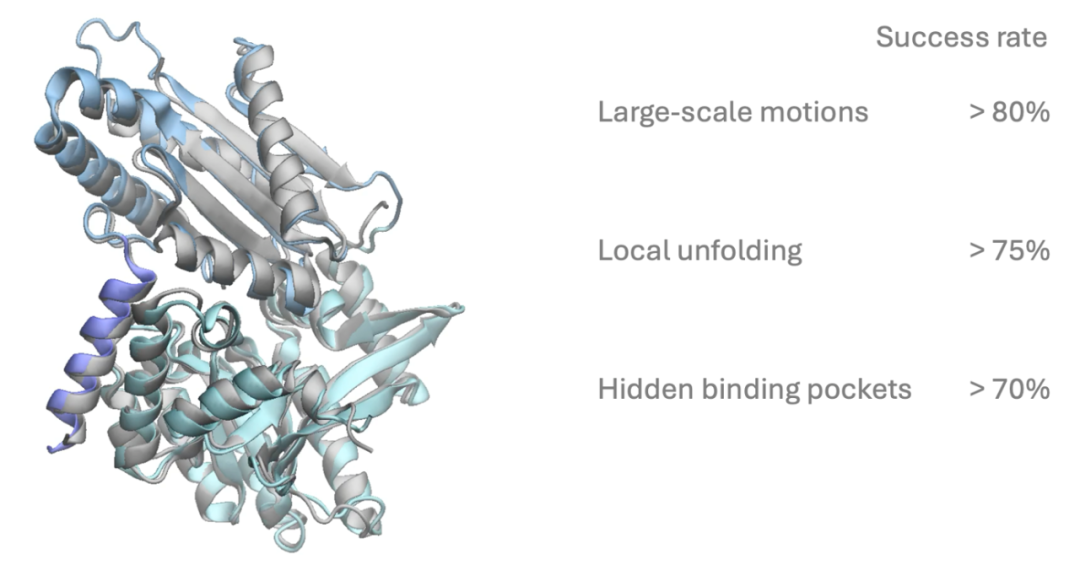
- Achieving a free energy prediction error of about 1 kcal/mol, highly consistent with millisecond MD simulations and experimental data, enabling several orders of magnitude acceleration over traditional molecular dynamics;
- Outstanding performance in predicting stability changes of mutants (ΔΔG), with mean absolute error below 1 kcal/mol and Spearman correlation above 0.6.
Open Source Release
The team has open-sourced the model parameters and code on GitHub and HuggingFace, along with over 100 milliseconds of MD simulation data covering thousands of proteins and mutants, providing rich resources for future research. BioEmu is also deployed on platforms like Azure AI Foundry and ColabFold, enabling easy access for users.
Future Outlook: From Single Proteins to Multi-Molecular Systems
The open-source release of BioEmu marks a significant step in promoting open science. The team is exploring extending BioEmu to more complex biological systems like protein complexes and protein-ligand interactions, integrating experimental data to improve model generalization and interpretability. In fields like structural biology, drug design, and synthetic biology, BioEmu is poised to become a bridge connecting structure and function, theory and experiment.