Chinese Renmin University Team Develops SPACE: A New Paradigm for DNA Foundation Models Based on MoE}
The SPACE model introduces a biologically inspired, MoE-based architecture for DNA modeling, achieving superior cross-species generalization and state-of-the-art performance in genomic tasks.


Author | Paper Team
Editor | ScienceAI
Sequence-to-function models are powerful genomic tools that predict functional features directly from DNA sequences, such as chromatin accessibility, gene expression, and splicing sites. Notable work includes Enformer [1], published in Nature Methods in 2021.
Recently, Google DeepMind released the AlphaGenome [2] model, which extended Enformer’s input sequence length to 1Mb, incorporated new training tasks like cleavage intensity and 3D DNA contact maps, and achieved single-base resolution predictions.
AlphaGenome has set new state-of-the-art (SOTA) performance across many genomic prediction tasks, reaffirming the potential of data scaling and the power of sequence-to-function supervised learning representations.
However, data expansion alone is not the only way to improve models. A research team from Renmin University of China’s Gaoling School of Artificial Intelligence proposed a systematic architectural innovation aligned with biological principles to enhance DNA foundation models more efficiently.
Their paper titled “SPACE: Your Genomic Profile Predictor is a Powerful DNA Foundation Model” was published on arXiv on June 2, 2025, and accepted at ICML 2025.

Link to paper: https://arxiv.org/abs/2506.01833
GitHub repository: https://github.com/ZhuJiwei111/space
Huggingface model: https://huggingface.co/yangyz1230/space
SPACE: Species-Profile Adaptive Collaborative Experts
Enformer and AlphaGenome face two intrinsic architectural limitations:
- 1. Limitations of shared species encoder: This “one-size-fits-all” encoder struggles to distinguish and model species-specific regulatory patterns and conserved evolutionary mechanisms.
- 2. Knowledge fragmentation in independent prediction heads: Genomic feature maps like chromatin accessibility, histone modifications, and transcription factor binding are highly interconnected. Separate heads cannot capture these dependencies, limiting deep understanding of complex gene regulation networks.

Figure 1: Overview of the SPACE model architecture. It includes three core stages: (1) CNN-based local context aggregation; (2) a species-aware MoE Transformer encoder; (3) an enhanced decoder with lineage grouping and dual gating mechanisms.
To address these challenges, the SPACE model introduces two key architectural innovations:
1. Species-aware Encoder: Incorporates sparse MoE layers within the Transformer encoder, consisting of a shared expert pool and multiple species-specific gating networks. For sequences from a particular species, the gating network dynamically selects and weights experts, enabling adaptive resource allocation and learning species-specific and shared regulatory features. An inter-information loss encourages specialization.
2. Lineage-Grouped Enhanced Decoder: Uses biological priors to group functionally related genomic maps (e.g., DNase-seq, ATAC-seq). A dual gating expert aggregation module dynamically selects relevant expert groups based on species and sequence information, modeling evolutionary conservation and dependencies among features. The refined predictions are fused with initial outputs via residual connections for stability and accuracy.
We conducted comprehensive evaluations on multiple genomic benchmarks.
Downstream Genomic Tasks
On the Nucleotide Transformer (NT) benchmark with 18 classification tasks, SPACE outperformed several leading models, including DNABERT-2, HyenaDNA, and NT series, achieving SOTA in 11 out of 18 tasks, demonstrating strong generalization and representation power.
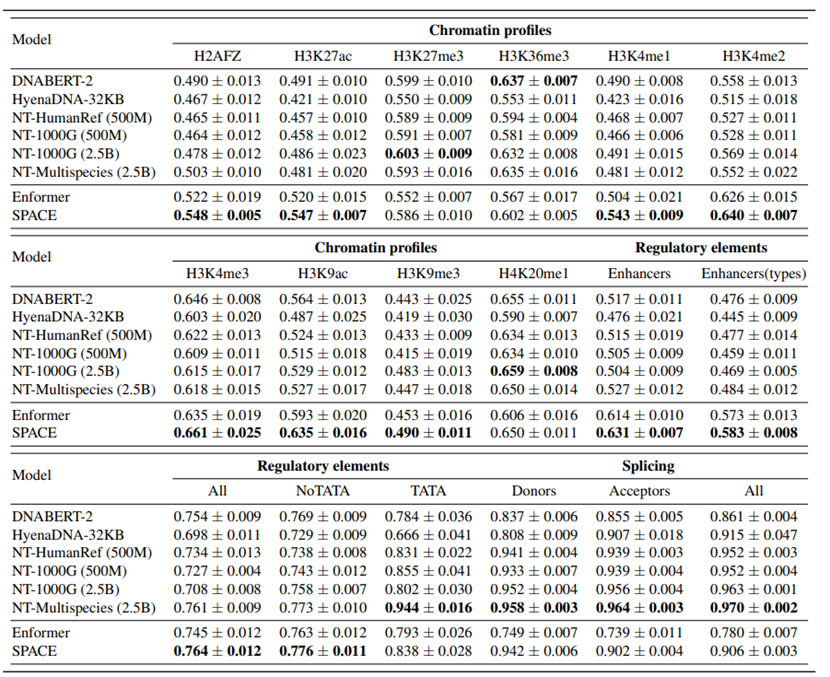
Figure 2: Performance comparison on NT downstream tasks. SPACE achieves top results in 11 tasks, outperforming unsupervised and supervised baseline models.
Cross-Species Generalization
To evaluate the impact of architectural improvements on cross-species generalization, tests were conducted on the GUE benchmark, including distant species like yeast (predicting epigenetic marks) and viruses (COVID variants). Compared to Enformer trained on human and mouse data, SPACE shows significant performance gains across all 11 tasks, especially a 27.28 percentage point improvement in yeast H3K4me3 mark prediction, confirming the effectiveness of the species-aware MoE encoder.
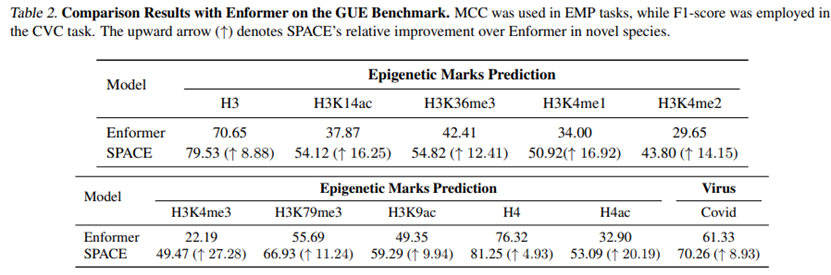
Figure 3: Cross-species generalization performance. SPACE significantly outperforms Enformer in yeast and virus prediction tasks, demonstrating robust transferability.
Summary
While AlphaGenome highlights the power of data scaling, our SPACE model offers an alternative path: introducing biologically aligned, dynamically adaptive architecture to deeply and efficiently capture complex regulatory language in DNA sequences. We believe this integration of architecture design and biological insights will pave the way for more powerful, interpretable genomic AI models. All code and models are open-sourced to promote community progress.
References:
- [1] Avsec, Žiga, et al. “Effective gene expression prediction from sequence by integrating long-range interactions.” Nature Methods 18.10 (2021): 1196-1203.
- [2] Avsec, Žiga et al. “AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model.” DeepMind (2025).