Chai-2 Achieves 100x Higher Accuracy Than Previous SOTA, Enabling Zero-Shot Antibody Discovery in 24-Well Plates}
Chai-2, supported by OpenAI, dramatically improves antibody design with 16% hit rate, over 100 times better than previous methods, enabling rapid zero-shot discovery in 24-well plates.


Editor | Luobo Pi
Designing functional antibodies reliably from scratch has always been a daunting challenge, requiring large-scale experimental screening of thousands or even millions of candidates to find targets effectively.
Supported by OpenAI, biotech company Chai Discovery introduced Chai-2, a multimodal generative model that achieved a 16% hit rate in completely de novo antibody design, over 100 times higher than previous computational methods.
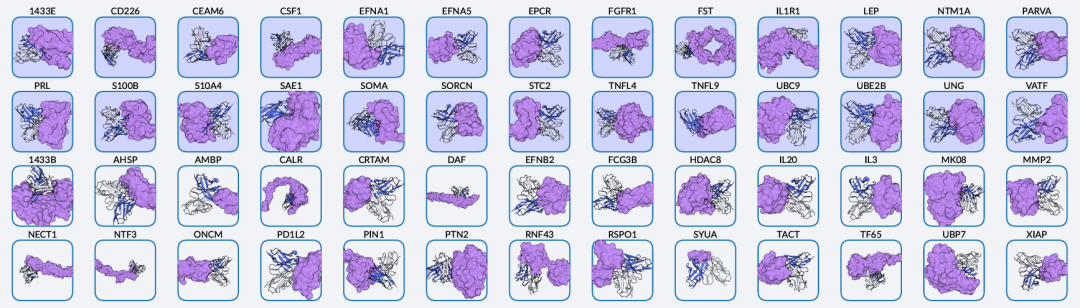
Illustration: Chai-2 targets 52 antigens. The blue box indicates that at least one successful binding was found in ≤20 tested designs for 50% of the tested targets. (Source: Paper)
Chai-2 integrates many advances from all-atom generative models. For example, the folding module predicts antibody-antigen complexes with twice the accuracy of Chai-1. It can generate candidate binding sites at any specific location, defined by just a few residues, entirely "zero-shot" without known starting sites.
Furthermore, Chai-2 can generate various sequence formats, including scFv antibodies, VHH domains, or miniature binding sites, and can bind multiple targets simultaneously, creating proteins with tailored cross-reactivity and selectivity. Importantly, all these functions require no target-specific adjustments, demonstrating Chai-2’s scalability across different applications.
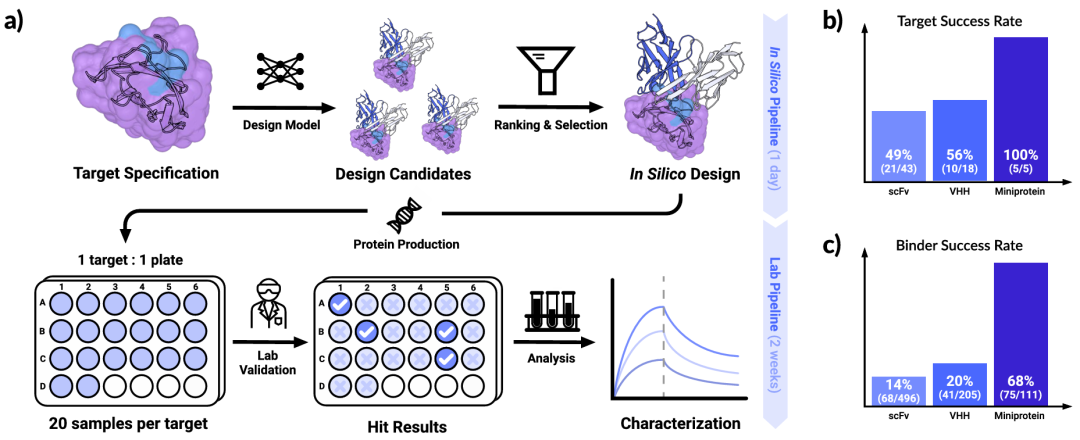
Illustration: Chai-2 reliably achieves state-of-the-art success rates in designing high-affinity proteins and antibodies. (Source: Paper)
Using prompt engineering, researchers designed over 50 types of binders for various targets, including mini proteins, VHH, and scFv, achieving top-tier experimental success rates across all categories.
They tested ≤20 antibodies or nanobodies per target, and at least one de novo binder was experimentally confirmed to bind 26 out of 52 new targets.
Unlike traditional biologics development, which relies on extensive and often undifferentiated experimental screening, Chai-2 employs a controllable, model-driven framework. Evaluating only 20 designs per target, researchers can identify potent binders within two weeks, potentially shortening development timelines from months or years to weeks.
Crucially, these targets are absent from existing protein databases, and with just one round of testing, they found at least 50% success rate for binding, with strong affinity and drug-like properties.
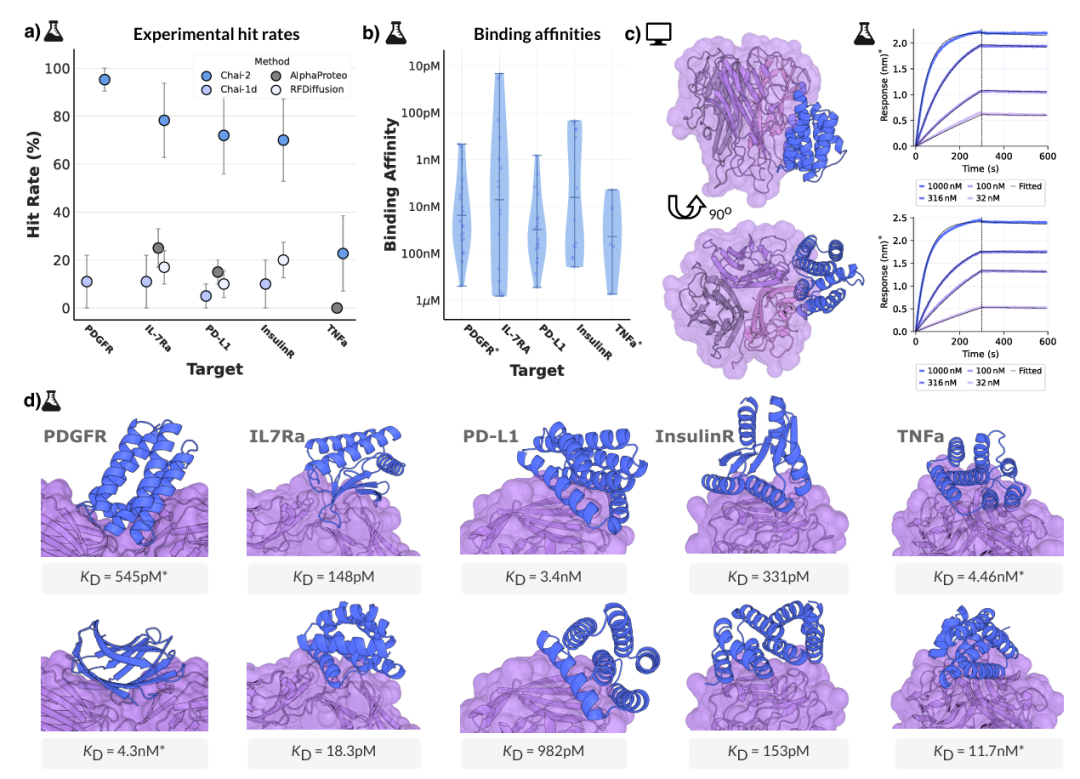
Illustration: Chai-2’s state-of-the-art performance in mini-protein design. (Source: Paper)
Beyond antibodies, Chai-2 achieved a 68% success rate in wet-lab experiments for mini-protein design, often generating binders with picomolar affinity. Its high success rate enables rapid validation and characterization within two weeks.
Its controllable, programmable design allows targeted exploration of molecular space, unlocking modes previously inaccessible to traditional methods.
Researchers can generate fully de novo designs in a single round, optimizing epitopes, scaffolds, and specificity constraints. This shift from random screening to targeted, programmable discovery suggests that even challenging antigens previously deemed undruggable could be tackled via computational design.
This approach could enable on-demand generation of epitope-specific binders, simplifying the development of advanced therapeutics such as antibody-drug conjugates, bispecifics, and multifunctional biologics. By atom-level reasoning—including ligand interactions and post-translational modifications—Chai-2 extends beyond traditional biologics to macrocycles, peptides, enzymes, and small molecules.
Looking ahead, the team envisions a phased pipeline for zero-shot candidate drug generation, where modeling of viscosity, pharmacokinetics, expression yield, and manufacturability can be optimized simultaneously across multiple key attributes.
"We believe our research will lead the shift from empirical discovery to deterministic molecular engineering. Combining atomic-resolution structure prediction with generative design, we break through traditional bottlenecks. Chai-2 marks a step toward our long-term vision of rational drug design: generating candidate drugs on the computer for IND submission in one go," the researchers stated in the paper.
Paper link: https://chaiassets.com/chai-2/paper/technical_report.pdf
Related content: https://x.com/chaidiscovery/status/1939684680447746050